Molecule Catalog
The molecule catalog is a list of molecules that are ready to be used on the platform. It can be accessed via the left navigation sidebar.
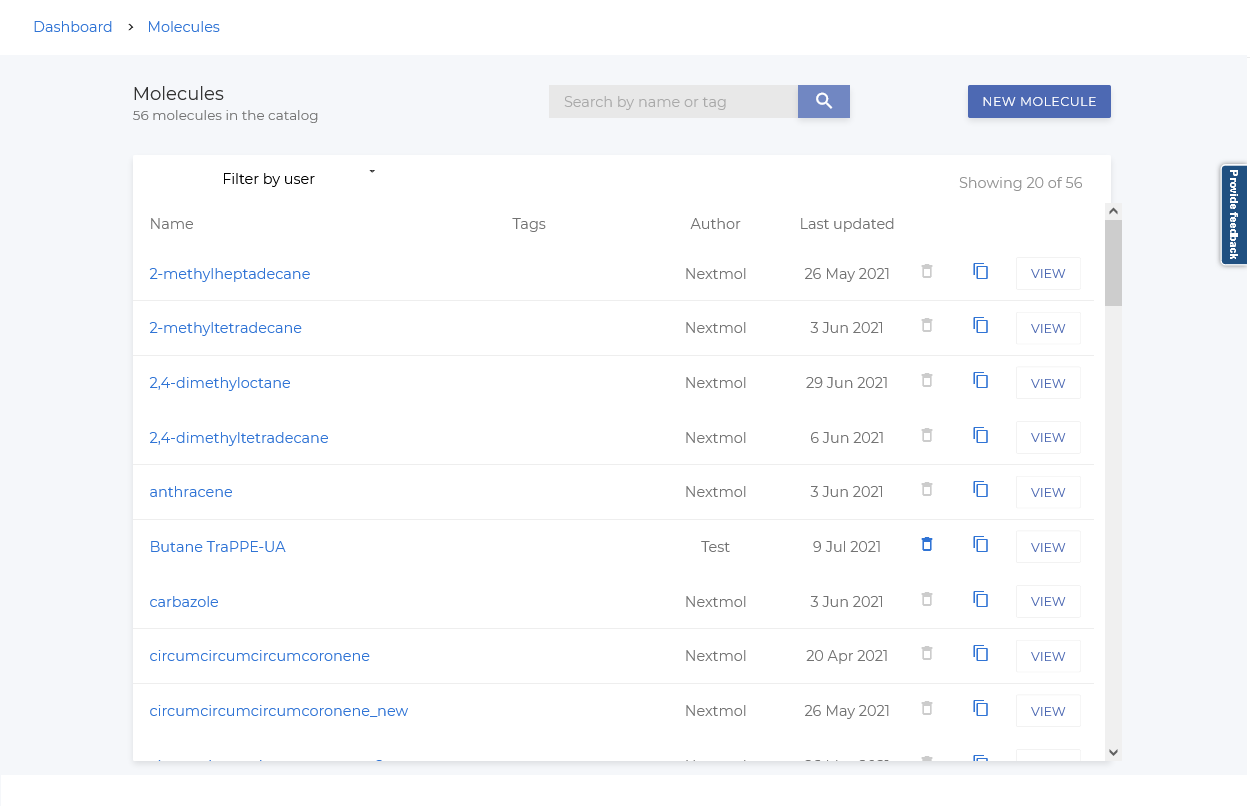
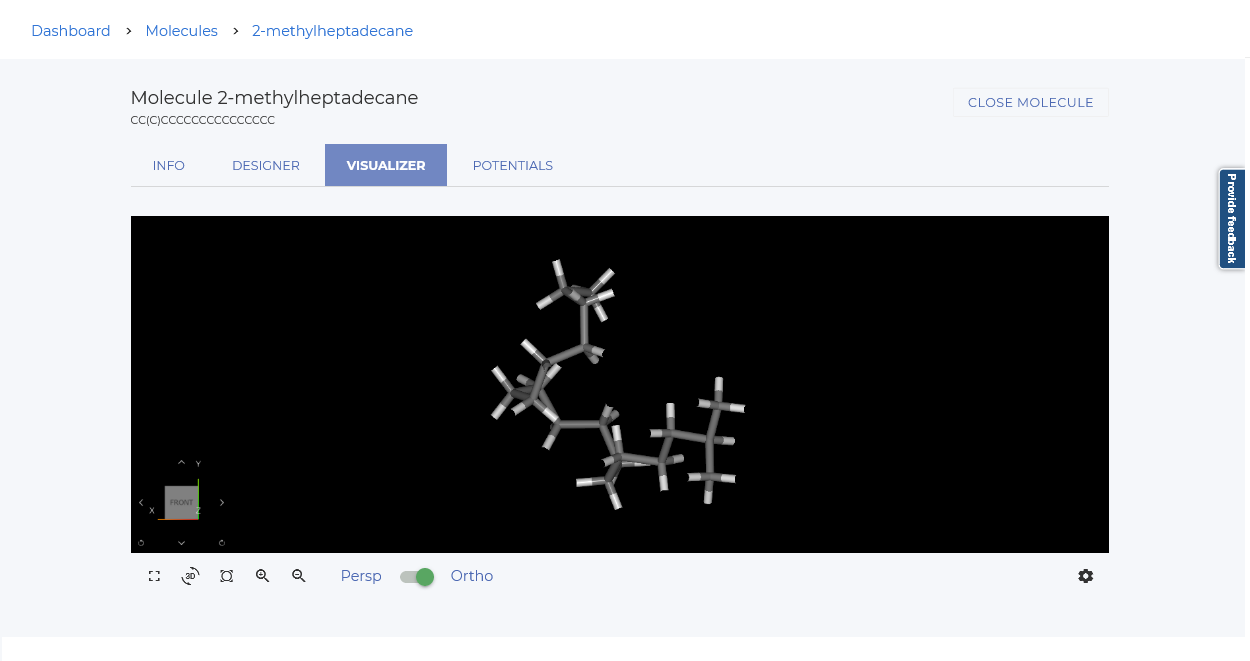
You can select a molecule from the catalog to visualize it.
It is also possible to create new molecules from the catalog by following these steps:
-
Click the NEW MOLECULE button.
-
Enter a name for the molecule.
-
Import a file containing the chemical structure (in PDB or GRO format) or enter the SMILES. When uploading a file, the platform checks for specific requirements: the file must not be corrupted (i.e., it should have a correct sequence of atom IDs, residue IDs, etc.), and no component should extend beyond the simulation cell (as this would prevent the unwrapping operation from succeeding). Molecules in PDB format without a cell definition are also allowed.
-
Save the molecule.
-
If no file or SMILES code has been provided, draw the 2D structure in the Designer tab. After sketching, click the
Save and convert to 3Dbutton. -
Create the topology in the POTENTIALS tab. To be used in a workflow, a molecule must be associated with a potential. The POTENTIALS tab allows you to either calculate a potential or upload a potential file. Currently, only gromacs '.top' files are supported for both creation and upload.
Once a molecule is in the catalog, you can either duplicate it () or delete it (
).