Building block catalog
The building block catalog is the place where building blocks are stored. The building blocks are the smallest units for the construction of the polymers. Two types of building block exist: the linear building blocks and the dendrimer building blocks.
Refer to the scientific introduction for a detailed description of the building block concept.
From the catalog, a user can also edit and create building blocks.
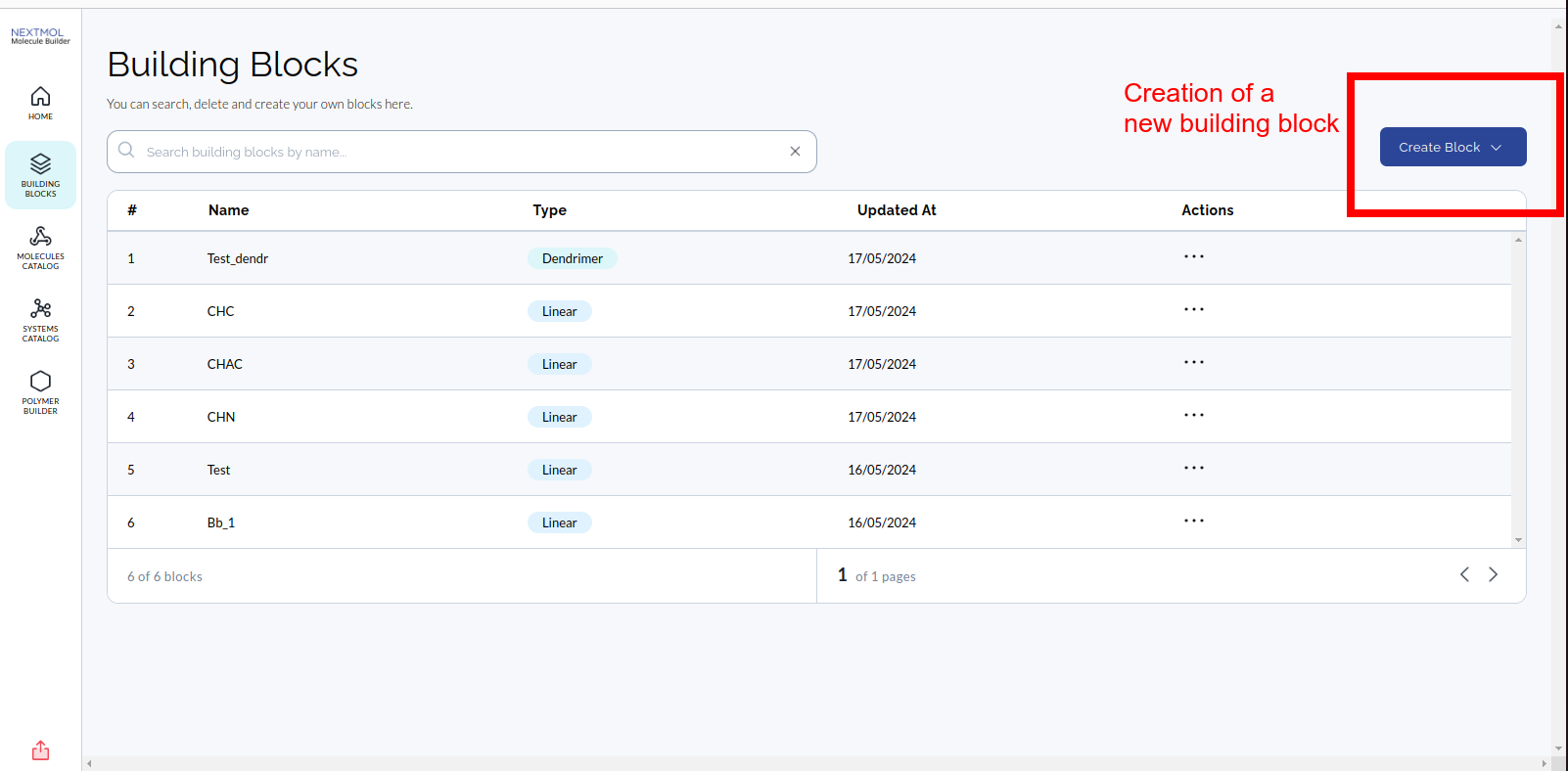
The searching bar on the top can be used to look by name, names are unique in the catalog, so a user can not create two building blocks with the same name. Every entry in the catalog can be opened and, from there, one can edit the elements that constitute the building blocks. The creation of a new building block should be done clicking on the blue botton on the top right of the page and selecting the type of building block to create (linear or dendrimer).
Creation and editing of building blocks
Once the type of building block to create is selected (clicking on the blue botton on the top right of the page) the building block creation page will be accessed. The definition page is different for linear and dendrimer building blocks, but both of them requires the definition of several molecular units. Two ways are available in the application for the definition of the molecular units: the direct use of SMILES or the use of the structure editor, a tool that allows to draw 2D molecular units. The two ways will be explained in the following sections. The same two ways can be used to edit an already saved building block.
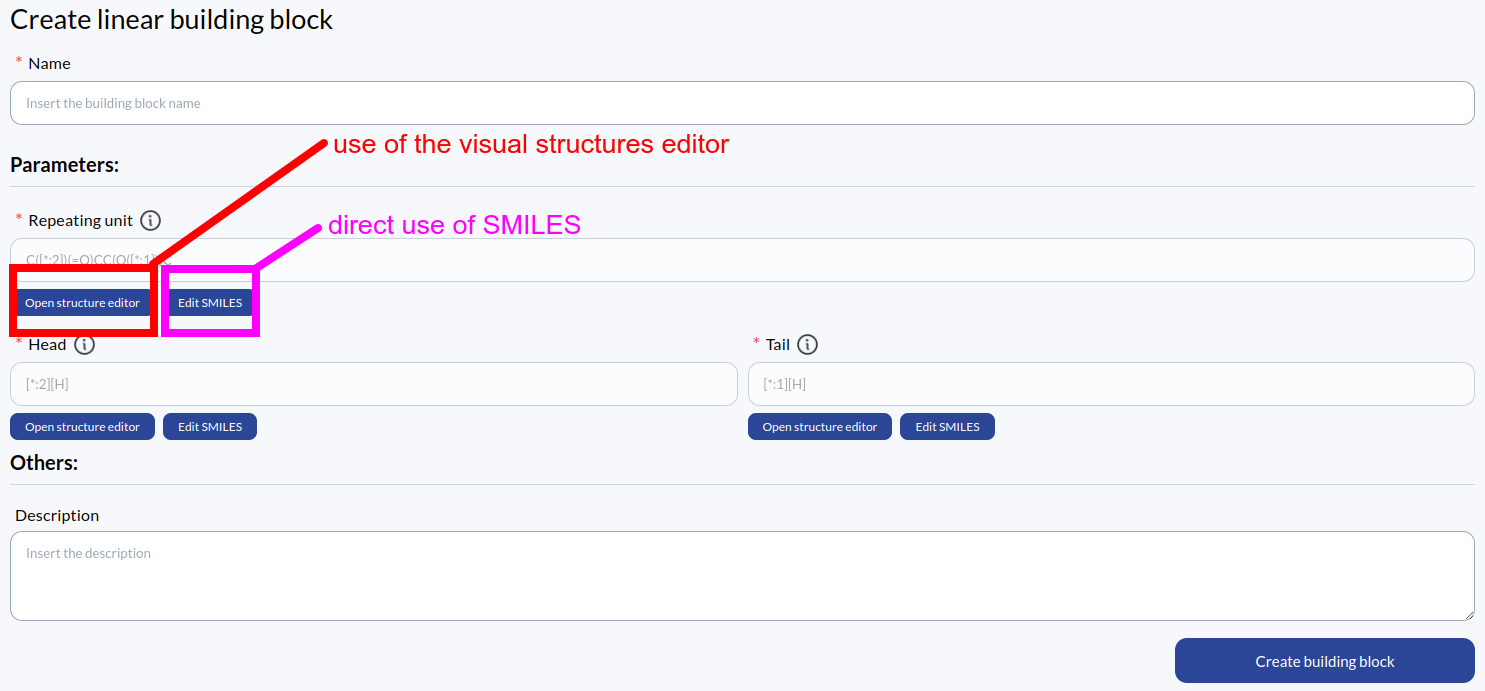
The direct use of SMILES
The user can type directly the SMILES of the molecular unit that must be created, remembering to follow the conventions on the open connections that are explaied in the introduction of the scientific language. In particular, the user should remember that the open connections must always be labelled with [*:1] if the connection will attach to the part of the polymer that is already built or [*:2] for the connections that are available for the next block in the construction. A [*:1] connection will always connect to a [*:2] connection.
The use of the structure editor
Integrated in our web application, we have a customized version of the the Ketcher, a molecule editor that allows to create 2D molecules. Here the user can draw the molecular units using all the Ketcher tools. The Save SMILES button will save the drawn molecule in SMILES form, which is the way we internally store molecular units. The structure editor is therefore useful to abstract away the concept of SMILES and allow the definition of molecular units without knowledge of the SMILES rules.
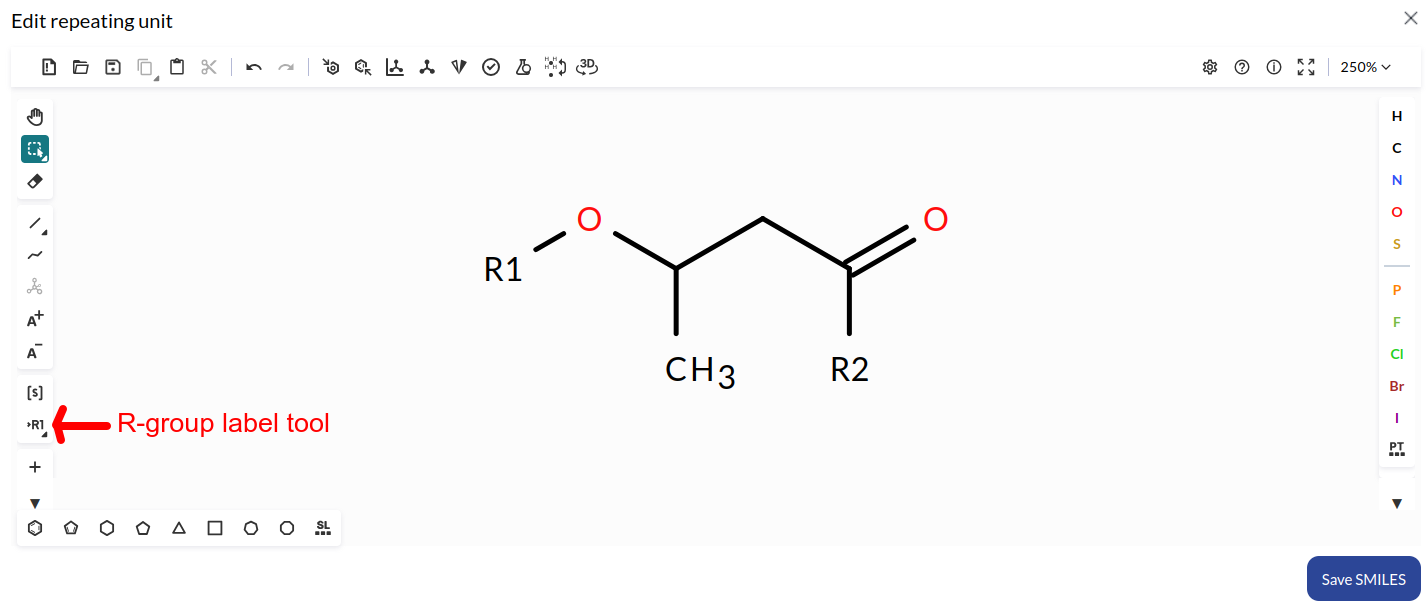
An important point to remember is that in the Ketcher (and therefore in our structure editor) the open connections must be created using the R1 and R2 labels (there is a R-group label tool on the left bar of the ketcher, see image above). The use of R1 will be translated to a [*:1] connection in the SMILES, the R2 will be translated into a [*:2].
Notes on the two types of building blocks
The definition of the linear building block requires 3 quantities:
-
Repeating Unit (REQUIRED): a molecular unit that contains one (and only one)
[*:2] (R2)connection and one (and only one)[*:1] (R1)connection. -
Head (OPTIONAL): a molecular unit containing one single
[*:2] (R2)connection, if not defined,[*:2][H]is used -
Tail (OPTIONAL): a molecular unit containing one single
[*:1] (R1)connection, if not defined,[*:1][H]is used.
The definition of the dendrimer building block requires 5 quantities:
-
Central Unit (REQUIRED): a molecular unit that contains one or more
[*:2] (R2)connections -
Repeating Unit (REQUIRED): a molecular unit that must contain one (an only one)
[*:1] (R1)connection and one or more[*:2] (R2)connections -
Capping Central Unit (OPTIONAL): a molecular unit containing one single
[*:1] (R1)connection, if not defined,[*:1][H]is used -
Head Repeating Unit (OPTIONAL): a molecular unit containing one single
[*:2] (R1)connection, if not defined,[*:2][H]is used -
Tail Repeating Unit (OPTIONAL): a molecular unit containing one single
[*:1] (R1)connection, if not defined,[*:1][H]is used