Tutorial 2: Creating a linear peptide
Description
Scope: this tutorial will guide the user through the creation of a linear peptide. We recommend the user to start this tutorial after finishing the Tutorial 1: Creating a linear polymer tutorial, as the generation of a peptide from scratch follows almost identical steps. It is important to note that currently the generation of a linear peptide yields a structure product of a random walk, i.e, a process that optimizes the structure after the addition of each aminoacid. Thus, the obtained structured will not be folded as in biological structures. Soonly, a new release of NEXTMOL Builder will provide this option.
Difficulty: easy
Time required: 5 minutes plus the computation time, which depends on the availability of the computational resources.
Creating a linear peptide segment
The peptide generation will follow the six-step procedure that was explained in Tutorial 1: Creating a linear polymer. However, the first step, i.e, the selection of the monomers, can be skipped as the standard aminoacids are preloaded in the platform. In the second step of the generation procedure, the user must click on 'Add linear peptide segment'. After indicating a name for the segment, the user must decide whether to use three or one letter codes for the declaration of the aminoacid sequence. In this case, we will use the three letter code, providing the sequence 'Glu-Glu-Met-Gln-Arg-Arg'. This sequence corresponds to 'Argireline', a popular peptide in products of the personal care sector.
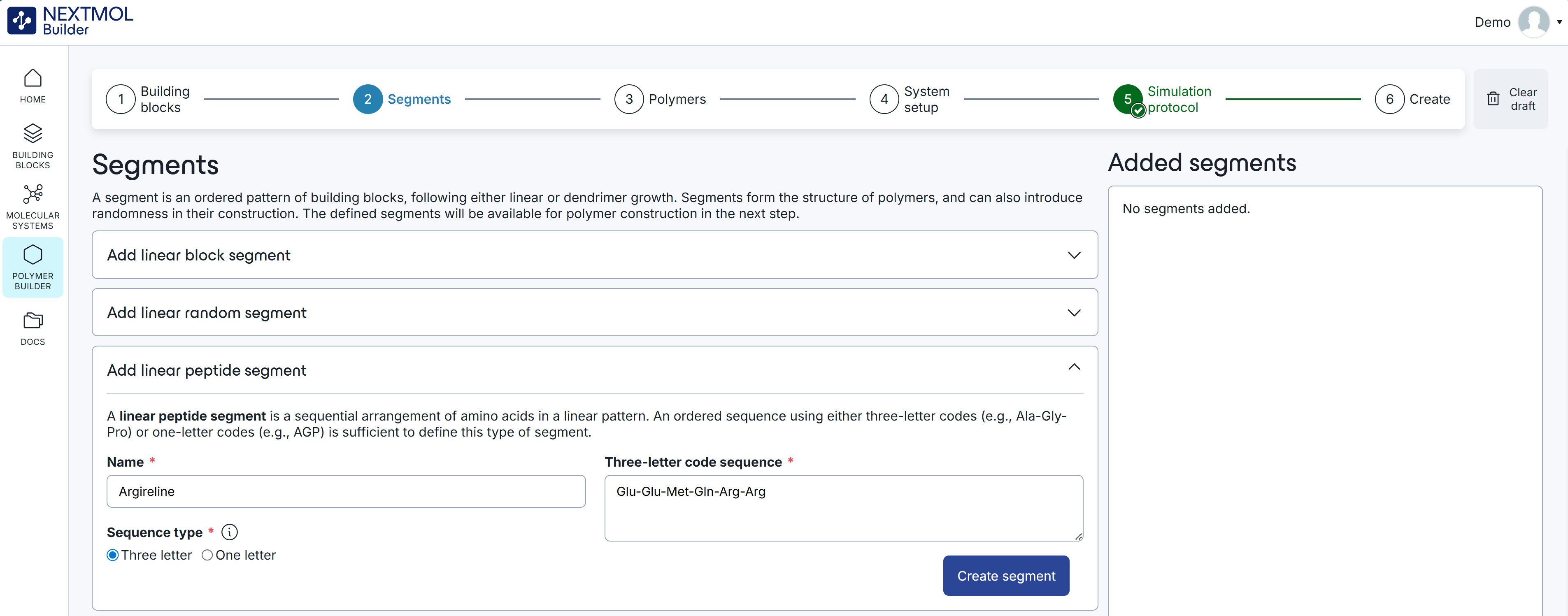
Figure 1: Declaration of the aminoacid sequence in a linear peptide segment.
Upon clicking on the 'Create segment' button, the segment will be saved in the 'Added segments' box. As many segments as desired can be saved, both peptide segments and polymeric segments.
Creating a peptide polymer
In the next step of the procedure the user can create the chains that will fill the simulation cell. After providing a polymer name (note that from a computational perspective chains are labelled as 'polymers'), the user must select the force field and the segments of the chain. The recommended force field for peptides is FF19SB, a go-to force field for peptides in commercial molecular dynamics packages. Then, the user can add the 'Argireline' segment that was created in the previous step.
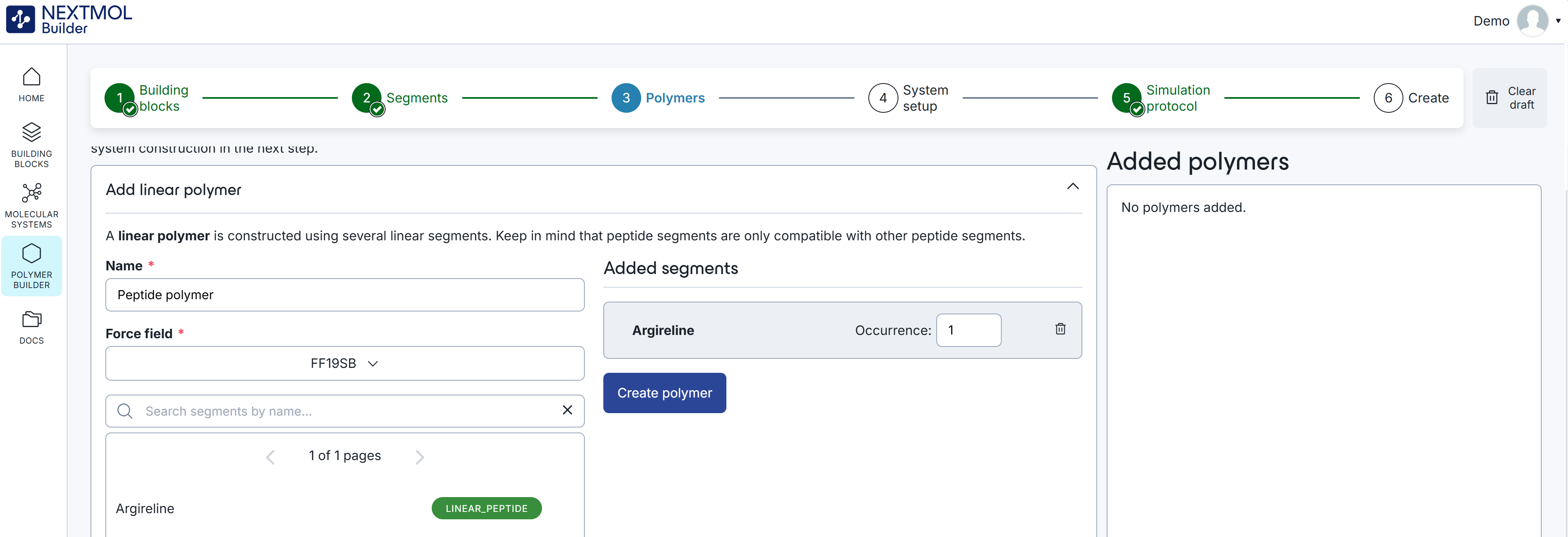
Figure 2: Creation of a peptide polymer.
Indicating the occurrence of the chain (in this case we will limit it to one) and clicking on 'create polymer' will save the result in the 'Added polymers' box. Note that several chains of different natures can coexist in a single cell, but mixed chains are not allowed. This means that once a linear peptide segment is added to a chain, the linear block and linear random polymeric segments will disappear from the list. The equivalent behavior will be observed if a polymeric segment is added first.
Last steps
The rest of the generation follows the same steps as any other polymer, as described in Tutorial 1: Creating a linear polymer. After defining the simulation cell, the setting of the minimization and dynamics and naming the system, which we called as the segment, 'Argireline', the generation can be run.
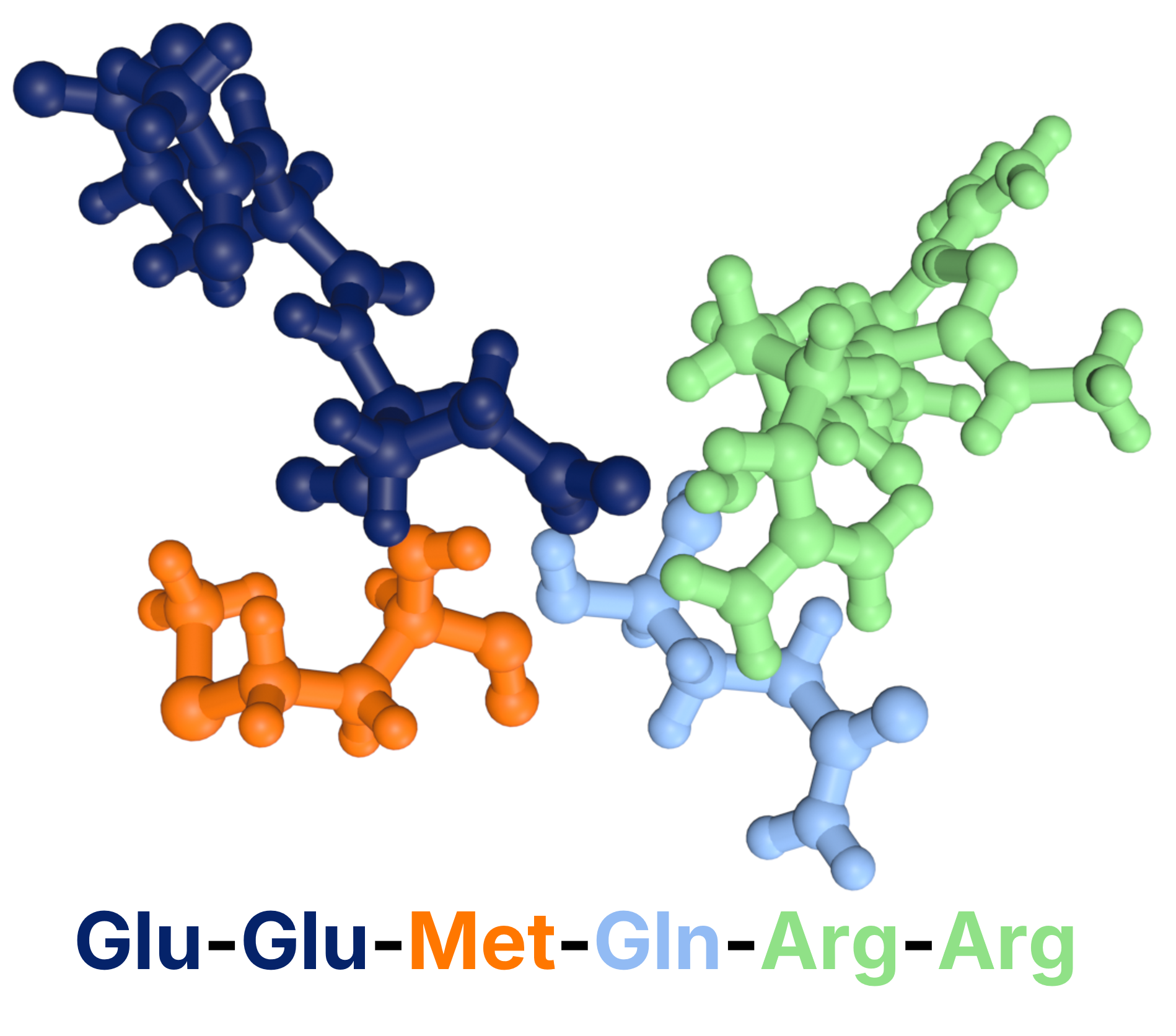
Figure 3: Argireline created with NEXTMOL Builder.